Coming Soon
- CITE-Seq Equivilent Method
- ATAC-Seq
The 10X Genomics Chromium Controller instrument is designed to rapidly automate the capture of thousands of single cells or DNA fragments, combining them with reagents in emulsion droplets to perform a variety of assays.
While some information is provided here on our site, we strongly recommend you obtain the latest methods, protocols and kit details from the support section of the 10X Genomics website.
Additional reagents; primers containing an Illumina read 1 sequencing primer, a 16 bp 10x Barcode, a 10 bp randomer and a poly-dT primer sequence are mixed with cell lysate and Master Mix. See figure below:

Upon dissolution of the Single Cell 3' Gel Bead in a GEM, primers containing (i) an Illumina R1 sequence (read 1 sequencing primer), (ii) a 16 bp 10x Barcode, (iii) a 10 bp randomer-UMI and (iv) a poly-dT primer sequence are released and mixed with cell lysate and Master Mix. Incubation of the GEMs then produces barcoded, full- length cDNA from poly-adenylated mRNA. After incubation, the GEMs are broken, and the pooled fractions are recovered. Libraries are generated and sequenced from the cDNA and the 10x Barcodes are used to associate individual reads back to the individual partitions. See figure below:

Sequencing parameters for 3'-RNA-Seq Libraries:
50,000 Read pairs per cell:
| Sequencing Read | Recommended Number of Cycles |
|---|---|
| Read 1 | 26 cycles |
| i7 Index | 8 cycles |
| i5 Index | 0 cycles |
| Read 2 | 98 cycles |
This solution offers the option to generate: 1) An enriched library from either T cells or B cells, directly from first-strand cDNA or 2) An enriched T cell library and/or an enriched B cell library, and/or a 5' gene expression library from amplified cDNA from the same cells.
The Single Cell V(D)J Solutions produce V(D)J enriched and 5' gene expression Illumina-ready sequencing libraries. A library comprises standard Illumina paired-end constructs which begin and end with P5 and P7. For V(D)J enriched libraries, Read-1 encodes the 16 bp 10x Barcode, 10 bp UMI, and 13 bp Switch Oligo, as well as the 5' end of an enriched transcript. For 5' gene expression libraries, Read-1 encodes the 16 bp 10x Barcode and 10 bp UMI. Due to Enzymatic Fragmentation, for both libraries Read-2 encodes a random internal fragment of the corresponding insert. Sample index sequences are incorporated as the i7 index read. See picture below:
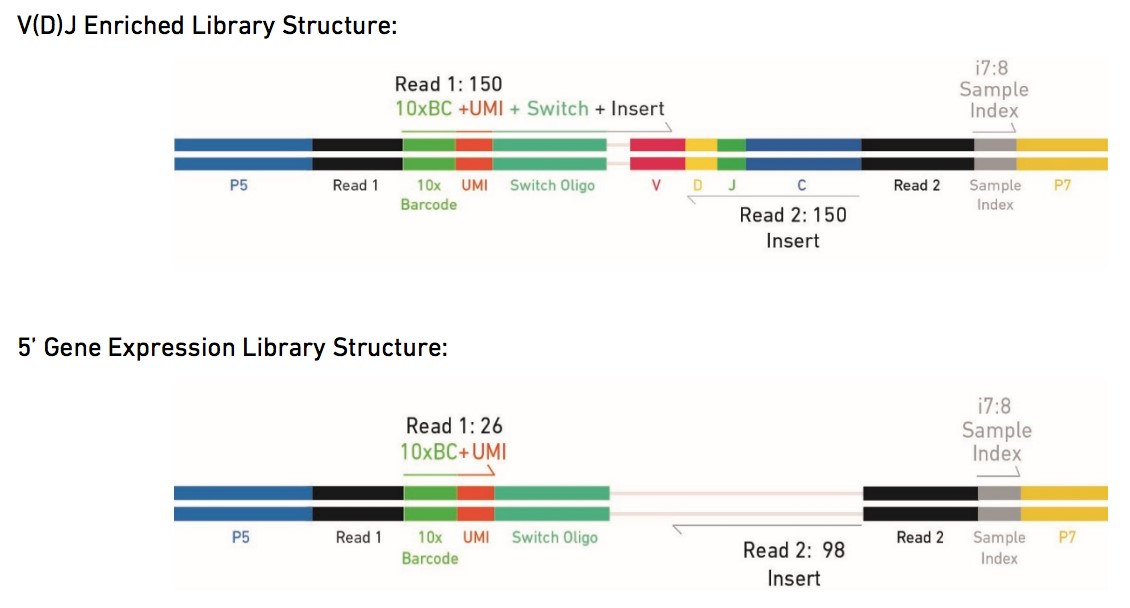
Sequencing parameters for V(D)J Enriched Libraries:
5,000 Read pairs per cell:
| Sequencing Read | Recommended Number of Cycles |
|---|---|
| Read 1 | 150 cycles |
| i7 Index | 8 cycles |
| i5 Index | 0 cycles |
| Read 2 | 150 cycles |
Sequencing parameters for 5' Gene Expression Libraries:
50,000 Read pairs per cell:
| Sequencing Read | Recommended Number of Cycles |
|---|---|
| Read 1 | 26 cycles |
| i7 Index | 8 cycles |
| i5 Index | 0 cycles |
| Read 2 | 98 cycles |
The 10X Chromium controller instrument is available for use in the BPF by trained users. Simply sign up on our calendar, acquire all your necessary reagents in advance, prepare your cells and execute your experiment.
The BPF does NOT provide full service processing of cells on the 10X Chromium instrument. We provide comprehensive training. To request training please email us at nextgen@genome.med.harvard.edu
The 10X Chromium controller is available for use 24 hours per day, 7 days per week. Once properly trained you will be given access to our 10X calendar to sign up to use the instrument. You will also be given card access to the lab where the 10X instrument is located.
Experiments are self service but a variety of cell handling recommendations can be found on the 10X website.
| Vendor | Service | Catalog Number | Cost |
|---|---|---|---|
| 10X | 10X Instrument use fee | 3873 | $168.90/hr |
| 10X/BPF | 10X Magnetic Separator (free rental use if returned to us within three business days): | 3667 | $590.32 |
The minimum time you can book is 2 hours; The time actually used does not impact price. The price is for each use rather than the amount of time. As such, we set bookings as 2hr blocks to protect your use time: in case you are running late, or you wish to use the thermal cycler next to the instrument, this avoids both scheduling conflicts and other issues.
Please contact us at nextgen@genome.med.harvard.edu if you have questions about pricing information for your specific project.
Please refer to the 10X website for the latest protocols and documents.
The BPF does not provide data analysis services but they may be available through the Harvard Chan Bioinformatics Core. Please contact them for more details. We do provide basic Cell Ranger output for 3'-RNA-Seq libraries only as well as bcl files.